Research Interests
Genome biology
Genome informatics
Single cell analysis
Systems biology
Speciation genetics
Research Projects
Our goal is to help understand the basis of how the genetic information is interpreted to enable proper development of various cell/tissue types particularly during embryogenesis. A combination of fluorescence time-lapse 3D imaging with automatic lineaging algorithm, we can automatically map gene expression with single cell resolution for every minute during C. elegans embryogenesis. In addition, we are able to produce precise information on migration and dividing axes of any cell during the development, which are critical for understanding how the different tissue types are formed.
Our research mainly focus on four broad themes , which are summarized below. More about our research can be found here.
- Application of cutting-edge genomic technologies in genome assembly, genome finishing and annotation
Existing genomes of various species have been produced mainly with Next Generation Sequencing (NGS) technology. The relatively short read length associated with NGS prevents generation of a high quality of genome assembly, especially in terms of continuity, leaving most genome drafts as fragmented pieces called contigs. This is mostly contributed by the presence of repetitive sequences. Third Generation Sequencing Technologies, including Single Molecule Real-Time (SMRT) Sequencing from PacBio and long-read sequencing from Oxford Nanopore Technologies, provide an opportunity for de novo genome assembly of high continuity, genome finishing and improvement in genome annotation. We have been appling these technologies mostly in C. elegans and its related species, C. briggsae to improve their existing genome assembly and annotation, which are used for subsequent comparison and functional characterization. See our publications for details.
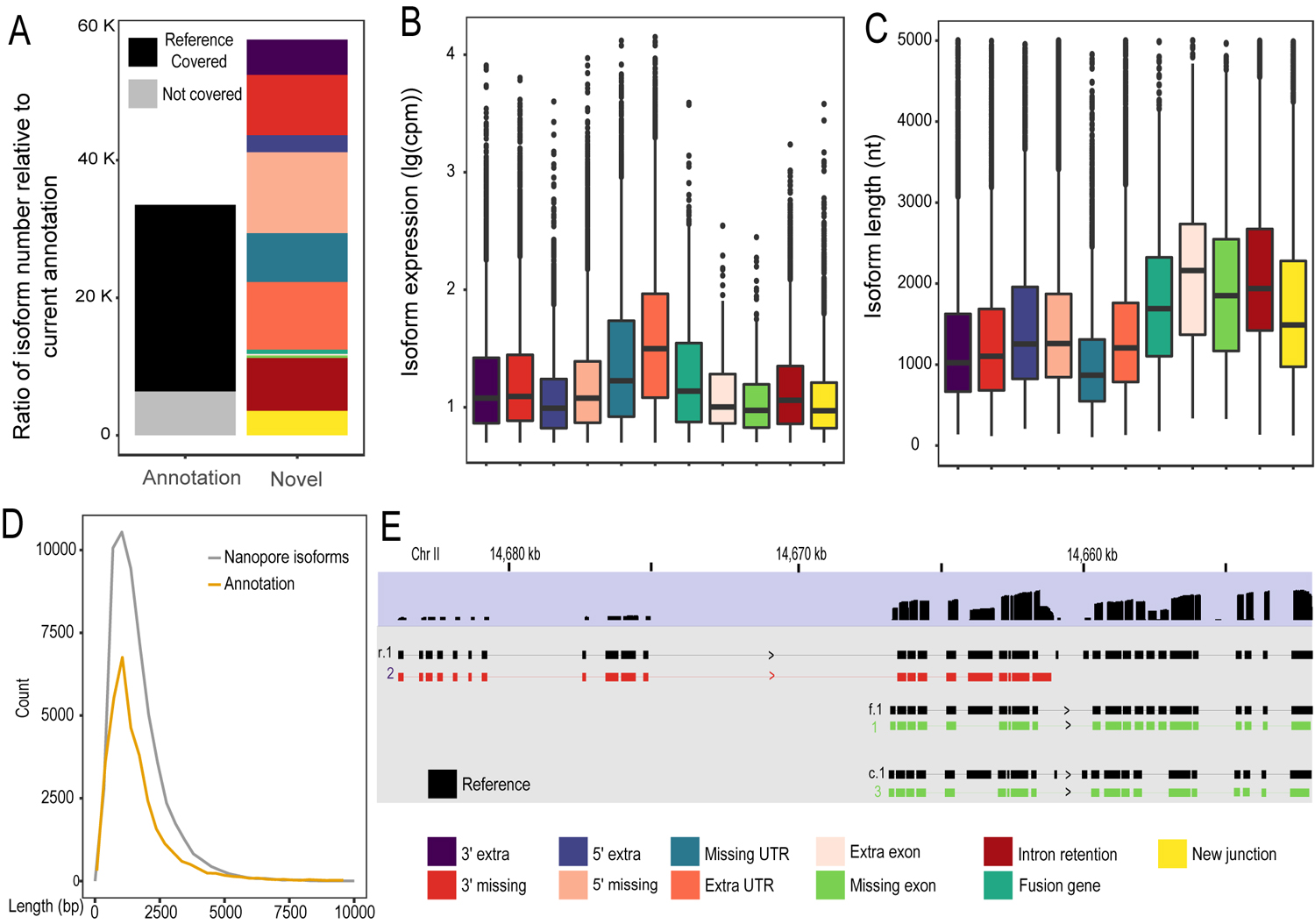
Newly called isoforms with Nanopore Direct RNA sequencing. (A) Summary of isoform calling using full-length long reads. Left: bar plots for the number of existing isoforms that are recovered or missed (grey) by the long reads. Right: bar plots for the number of novel isoforms called by TrackCluster. (B-C) Expression level (B) and length distribution (C) of the novel isoforms of the various categories. (D) Count distribution of existing isoforms or the isoforms output by TrackCluster over their length. (E) Example of TrackCluster predicted isoforms.
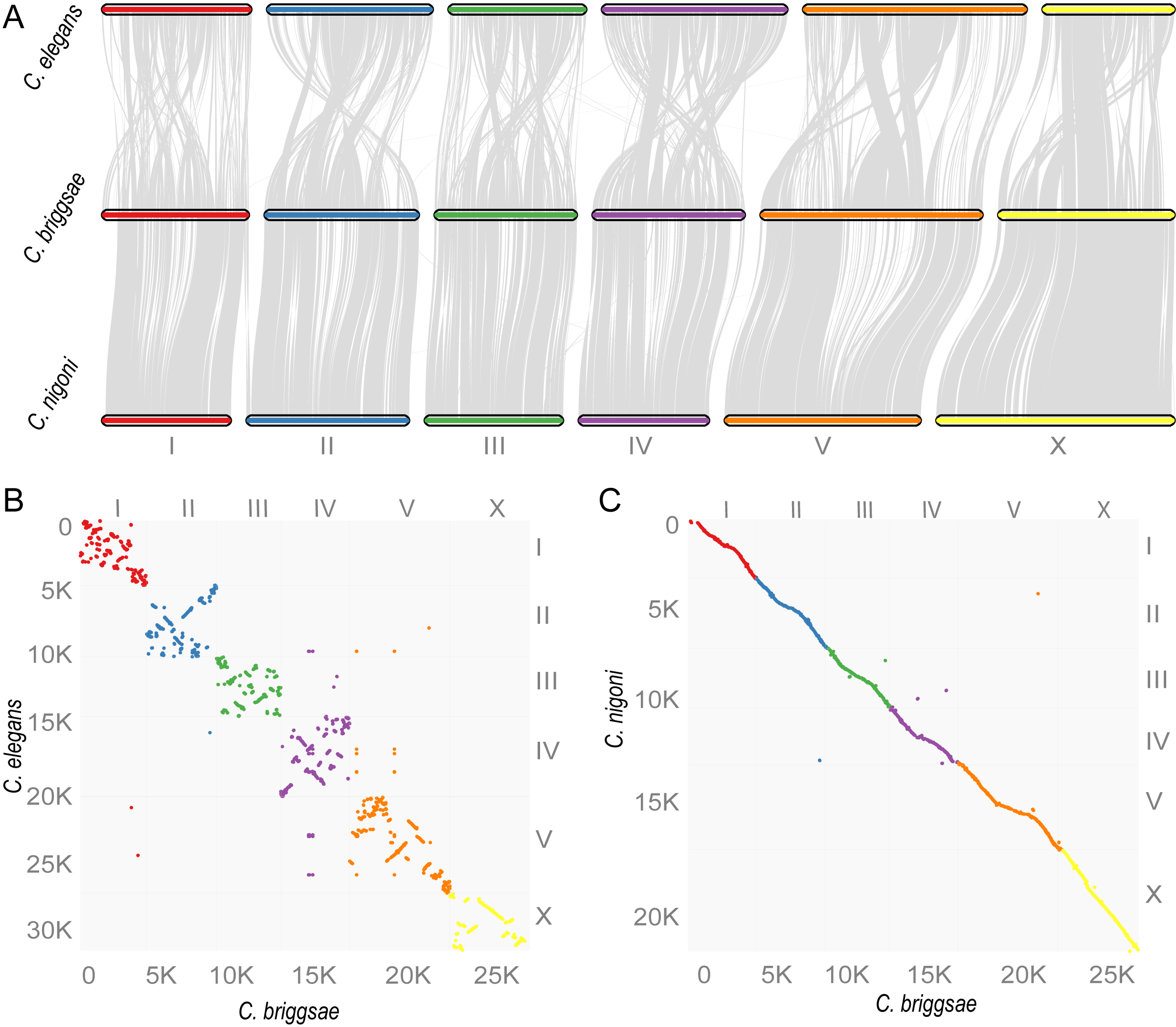
Comparison of synteny consisting of gene blocks between C. briggsae and C. nigoni or C. elegans. A new C. nigoni genome assembly was generated with Nanopore long reads, Illumina Synthetic long reads and the reads of mate pair sequencing of BAC and fosmid ends. A total of 14,427 and 7,679 orthologous pairs were used for comparison between C. briggsae and C. nigoni or C. elegans, respectively, with a bin size of 30 genes. (A) Syntenic view. (B-C) Dotplot view.
- Mechanism of postzygotic hybrid incompatibilities between nematode species
Hybrid incompatibility (HI), including sterility and lethality between closely related species has been noticed over a century ago, but only recently is it possible to decipher the molecular basis of speciation. Genetic approaches play a central role in speciation research. However, genetic study on speciation reaches its bottleneck due to the insufficient genetic tools available to the species of interest. Nearly all of the model organisms have been used for speciation research due to the availability of abundant molecular and genetic tools. However, C. elegans has not been explored for such research because there is no sister species found with which it mates and produce hybrid viable progeny. C. briggsae is the closest living relative of C. elegans with sequenced genome. A new species, sp.9 that can mate and produce a few viable hybrid progeny with C. briggsae has recently been isolated by Marie Felix and colleagues, opening the door for the speciation research using the species pair. We try to develop new tools and resources to make C. briggsae as an attractive model system for research into molecular mechanisms of hybrid incompatibility.Developmental dynamics of signaling pathways during nematode embryogenesis
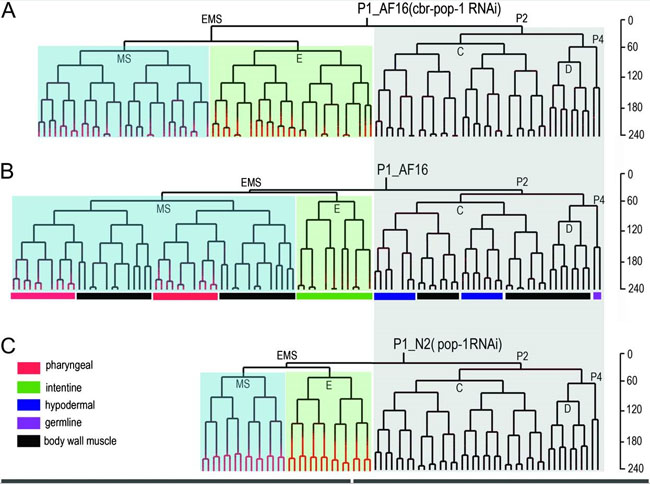
Signal transduction pathways such as Wnt and Notch pathways play key roles in diversifying cell types during the development of multicellular organisms. The two pathways are superficially conserved at the protein sequence level. However, the functional conservation of the pathways may not parallel to that of sequences. One surprising example is the opposite lineage transformation in EMS between C. elegans and its close relative, C. briggsae after depletion of Wnt pathway activities (See figure below). We have developed genetic tools which allow adaptation of automatic lineaging in C. briggsae. Parallel lineaging of the two species coupled with tissue specific fluorescent markers will facilitate the identification of further functional dynamics between the species pair.
Differential regulation of EMS lineage transformation by Wnt signaling pathway
- Gene regulatory networks underlying cell fate determination during nematode embryogenesis
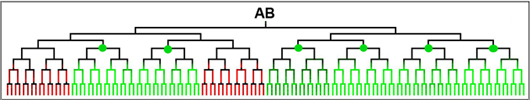
During C. elegans embryogenesis, a total of 558 cells are produced from a fertilized egg within about 14 hours. Majority of these cells are differentiated into its final cell fates before hatching. The developmental potential of the fertilized egg is restricted in a stepwise manner following each cell division by differentially expressing a cohort of regulatory proteins. Together with the genetic and genomic tools, especially CHIP-Seq, our unusually high temporal and spatial resolution of gene expression patterns derived from automatic lineaging provides a unique opportunity to dissect the gene regulatory network underlying the tissue/organ formation (see figure below).
Lineally complementary expression patterns suggest a negative regulatory relationship between the gene pairMajor Research Equipment
Leica Sp5 confocal microscope equipped with two HyD detectors and water immersion objective len
Brand new Leica Stellaris confocal microscope equipped with two HyD detectors and water immersion objective len
Competitive Research Grants from Hong Kong Research Grant Council (RGC)
1. Principle coordinator (PC) of Collaborative Research Fund (CRF) awarded by Hong Kong Research Grant Council (RGC), 2012-2015
2. PI of General Research Fund (GRF) funded by Hong Kong RGC, 2023-2026
3. PC of Innovation and Technology Fund (ITF) funded by Hong Kong ITC, 2023-2025
4. PI of General Research Fund (GRF) funded by Hong Kong RGC, 2022-2025
5. PI of General Research Fund (GRF) funded by Hong Kong RGC, 2020-2023
6. PI of NSFC/RGC Joint Research Scheme funded by Hong Kong RGC and NSFC, China, 2018-2022
7. PI of General Research Fund (GRF) funded by Hong Kong RGC, 2018-2021
8. PI of General Research Fund (GRF) funded by Hong Kong RGC, 2017-2020
9. PI of General Research Fund (GRF) funded by Hong Kong RGC, 2016-2019
10. PI of General Research Fund (GRF) funded by Hong Kong RGC, 2015-2018
11. PI of General Research Fund (GRF) funded by Hong Kong RGC, 2014-2017
12. PI of Early Career Scheme (ECS) Award funded by Hong Kong RGC, 2012-2016
13. Co-PI of Collaborative Research Fund (CRF) awarded by Hong Kong Research Grant Council, 2018-2021
14. Co-PI of Collaborative Research Fund (CRF) awarded by Hong Kong Research Grant Council, 2015-2018
15. Co-PI of GRF funded by Hong Kong RGC, 2020-2023
16. Co-PI of GRF funded by Hong Kong RGC, 2018-2021
Professional services
02/2013 - 01/2019, panel member of Hong Kong RGC, Biology and Medicine Panel, Hong Kong RGC and NSF China joint Scheme
01/2019 - present, Grant reviewer for NSF, USA
10/2018 – present, Grant reviewer for NSERC, Canada
11/2021 – present, Editorial Board of BMC Biology
11/2020 – present, Editorial Board of Frontiers in Physiology
11/2016 – present, Editorial Board of Scientific Reports
06/2005 - present, reviewer for journals Genome Research, PLoS Genetics, Nucleic Acid Research, Genetics, Genome Biology, GigaScience, G3 among others